15-Jan-2021
Single particle Cryo-EM has evolved extremely rapidly in the recent years (with great advances in the quality of the instrumentation and the methods used for data analysis) and has become the technique of choice to determine the structure of membrane proteins or other macromolecular complexes that are challenging to crystallise. However, despite the amazing advances in this field of structural biology, several factors such as particle size and the non-random orientation of particles in ice can strongly affect the resolution of 3D reconstructions when using Cryo-EM which makes the study of small particles or strongly oriented molecules extremely difficult.
In order to overcome these limitations, Tomasz Uchanski and colleagues from the VUB-VIB Instruct Centre in Belgium have designed new chimeric molecules called megabodies that increase the molecular weight of target molecules and can overcome the preferential orientation of particles in ice resulting in better reconstructions when using single particle Cryo-EM.
These megabodies are composed of a single-domain antigen binding protein such as a nanobody or a monobody that recognises their target protein with high affinity, grafted into a larger scaffold protein via two peptide linkers. The association of nanobody-like proteins, that are known to stabilise their target protein complexes and reduce the conformational heterogeneity, with a larger scaffold results in megabodies that are conformationally stable and that can, unlike nanobodies alone, overcome preferential orientation at the water-air interface.
Megabodies can be assembled using different combinations of scaffold proteins and antigen binding proteins and the construction of megabodies with new specific nanobodies can easily be achieved which makes this technique accessible to all. The principle of deriving megabody libraries from nanobody-immune libraries was tested successfully using a nanobody library from a GFP-immunised llama where functional megabodies that are properly expressed and that recognise their target antigen can then easily be selected in phage or in yeast cells.
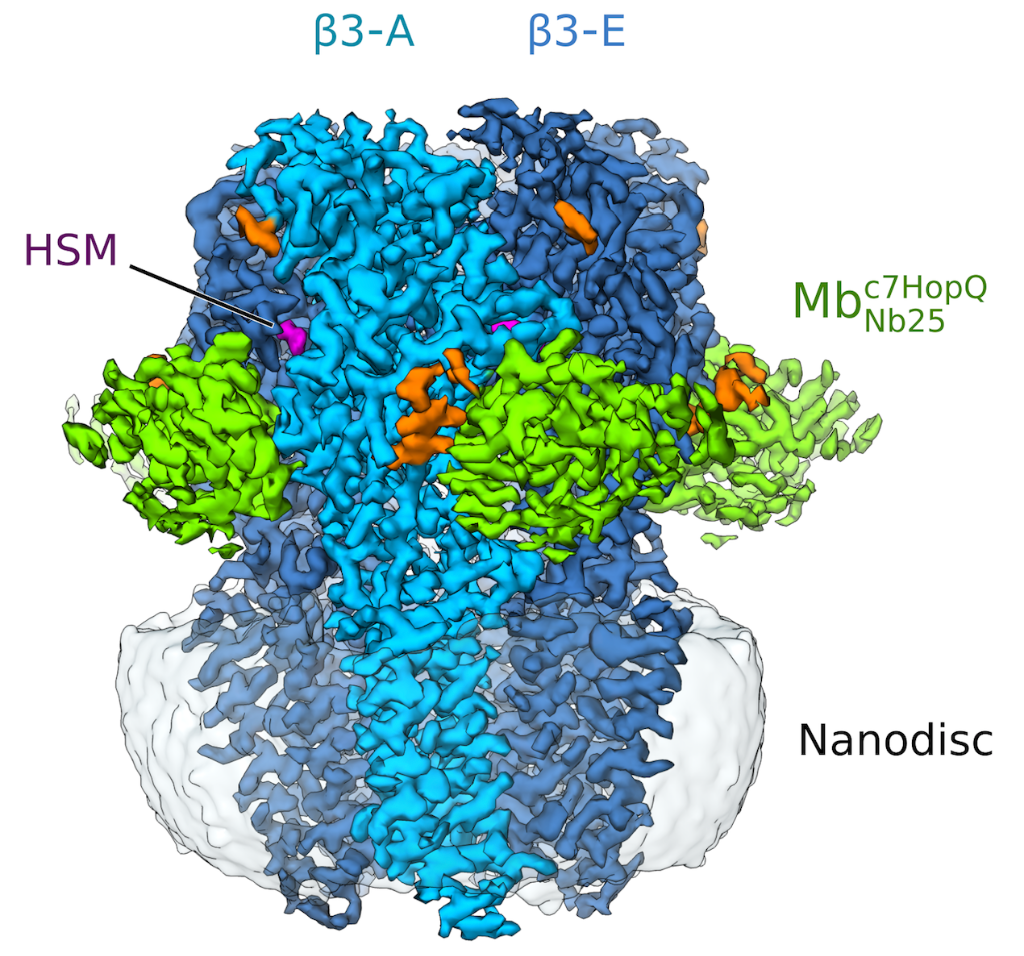
Figure 1: Side view of the sharpened Cryo-EM density map of histamine-bound homopentameric GABAA- b3 receptor in a complex with the megabody Mbc7HopQNb25 in lipid nanodisc. Five b3 subunits are coloured light blue (subunit A) and dark blue (subunits B-E). Histamine (HSM), Mbc7HopQ and glycans are coloured magenta, green and orange respectively.
The utility of megabodies in single particle Cryo-EM was tested on the structural analysis of the human GABAA receptors that are known to display a strong preferential orientation in ice making the study of these membrane protein structures extremely challenging.
Remarkably, when bound to specific megabodies, both homomeric and heteromeric GABAA receptors displayed a more randomised orientation in ice which greatly improved the 3D reconstruction of receptors in lipid bilayers using single-particle Cryo-EM.
Thanks to the use of a megabody recognising GABAAR-β3 subunit, the structure of the GABAAR-β3 homopentamer was resolved at 2.49 Å resolution (see figure 1), allowing a clear view of the small agonist histamine (figure 2).
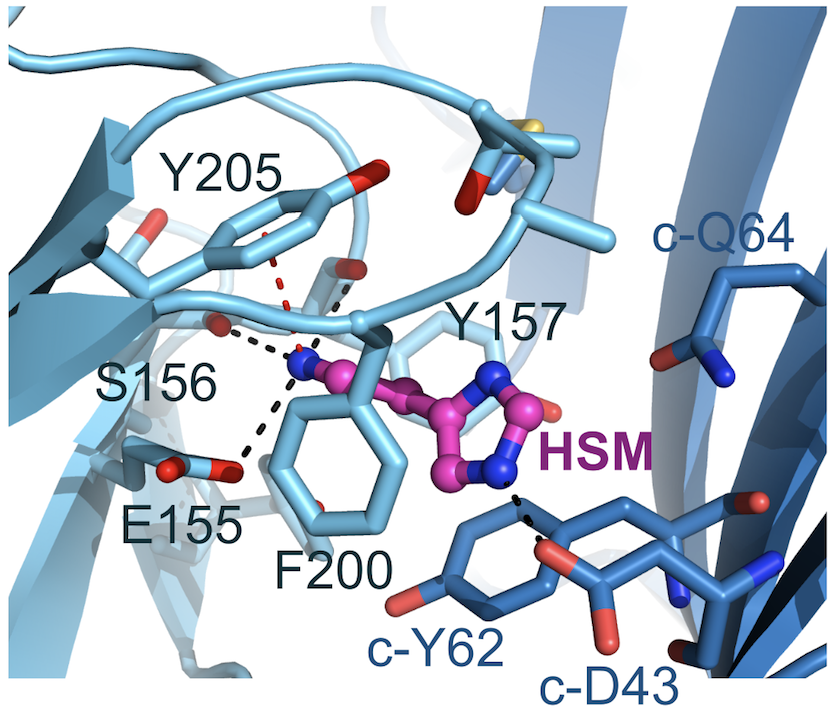
Figure 2: Histamine binding mode. Complementary face residues from E subunit marked by ‘c-’. Black and red dashed lines indicate putative hydrogen bonds and cation-π interactions, respectively.
Read full text of the article Uchański, T., Masiulis, S., Fischer, B. et al. Megabodies expand the nanobody toolkit for protein structure determination by single-particle cryo-EM. Nat Methods 18, 60–68 (2021). https://doi.org/10.1038/s41592-020-01001-6
Read more about nanobody discovery at Instruct-ERIC.
Read more about CryoEM at Instruct-ERIC.