23-Aug-2019
Bacteriophages are viruses that infect and replicate within bacteria and archaea. Recent work at the Instruct-ES Centre (CNB-CSIC) in collaboration with the IRB-CSIC centre in Barcelona, has elucidated critical aspects of the T7 tail protein structure and mechanism, providing crucial insights into how phages infect bacteria. Such information could prove invaluable in the fight against antimicrobial resistance, and may also lead to novel applications in the use of phages as therapeutic agents to deliver genes to target cells.
T7 is a double-stranded DNA virus within the Podoviridae family of phages, which infects susceptible bacterial cells, including most strains of E. coli. Double-stranded DNA bacteriophages package their genome at high pressure using a set of tail proteins to seal the pressurised DNA. Interaction with the membrane of the bacterial host triggers a phage portal to open, allowing it to infect the host by injection of DNA.
Combining X-ray crystallography with high resolution cryo-electron microscopy and medium resolution cryo-EM to solve the X-ray phases, Ana Cuervo, Moserrat Fàbrega-Ferrer and colleagues have visualised the portal structure of T7 bacteriophage isolated and in complex with the other tail proteins. The overall shape of the T7 portal protein is a ring-like structure, made up of 12 subunits with an axial central channel. The tail machine comprises the portal (gp8), the adaptor (gp11), the nozzle (gp12) and the fibres (gp17), and the study provides a model for the roles of each of the fibre-less tail proteins during T7 bacteriophage packing, retention and ejection.
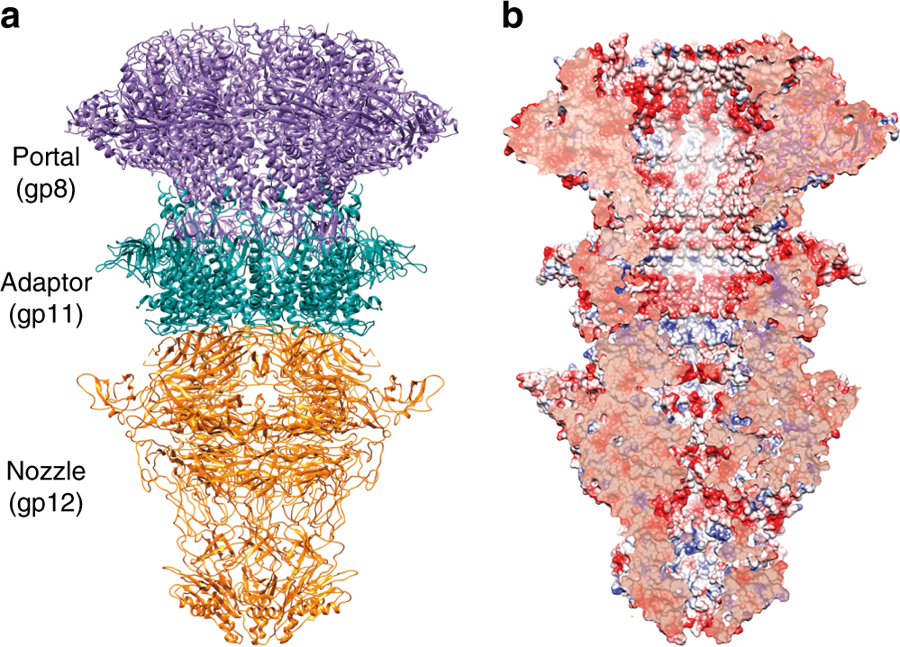
a. Ribbon representation of the cryo-EM T7 tail structure (gp8-gp11-gp12). Gp8 portal, gp11 adaptor, and gp12 nozzle proteins are shown in purple, green, and orange, respectively. b. Longitudinal cut of the electrostatic potential surface.
The portal protein gp8 was found to exist in two conformations, relating to the open and closed state. Interestingly, the mechanism for the transition between the two states was shown to be a movement at the crown (6A) and the opening of a hinge domain. This movement is illustrated in the animation below.
Another unexpected finding was that the gp12 tail nozzle shows a fold with six beta-propellers, which are now known to be essential to tightly close the channel gate in the mature phage. This fold shows no structural similarity with any bacteriophage tail previously reported. There are four closing gates with negatively charged residues found along the gp12 internal channel, with apertures ranging from 8.6A to 23A. It is proposed that the negative charge is essential to avoid the DNA sticking to the channel during ejection.
These studies demonstrate the power of integrative structural biology, with X-ray crystallography and Cryo-EM used complementarily to solve challenging molecular structures.
Read more about Cryo-EM, X-ray crystallography, crystallisation and image processing services at Instruct-ERIC.
Read the full text article: Cuervo A., Fàbrega-Ferrer M. et al. 2019. Structures of T7 bacteriophage portal and tail suggest a viral DNA retention and ejection mechanism. Nature Communications 10: 3746.