All publications from iNEXT-Discovery acknowledge the project and are appearing as Open Access!
< 21-40
|
20 |
Mar 2, 2021 |
Mixed-valence compounds as polarizing agents for Overhauser dynamic nuclear polarization in solids doi: 10.1002/anie.202103215 Gurinov A, Sieland B, Kuzhelev A, Elgabarty H, Kühne TD, Prisner T, Paradies J, Baldus M, Ivanov KL, Pylaeva S. Angew Chem Int Ed Engl. 2021 Jul 5;60(28):15371-15375 |
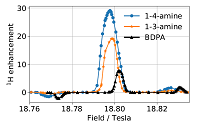 |
|
19 |
Feb 12, 2021 |
Protein in‑cell NMR spectroscopy at 1.2 GHz doi: 10.1007/s10858-021-00358-w Luchinat E, Barbieri L, Cremonini M, Banci L. J Biomol NMR. 2021, 75:97-107 |
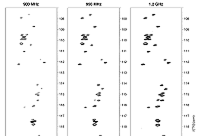 |
|
18 |
Feb 10, 2021 |
Real-time NMR spectroscopy in the study of biomolecular kinetics and dynamics doi: 10.5194/mr-2021-16 Pintér G, Hohmann KF, Grün JT, Wirmer-Bartoschek J, Glaubitz C, Fürtig B, Schwalbe H. Magn. Res. Disc. 2021 |
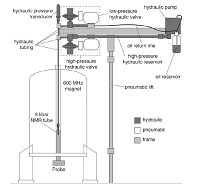 |
|
17 |
Feb 4, 2021 |
The folding landscapes of human telomeric RNA and DNA Gquadruplexes are markedly different doi: 10.1002/anie.202100280 Muller D, Bessi I, Richter C, Schwalbe H. Angew. Chem. Int. Ed. 2021, 10.1002/anie.202100280 |
 |
|
16 |
Dec 20, 2020 |
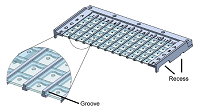
Facilitated crystal handling using a simple device for evaporation reduction in microtiter plates doi: 10.1107/S1600576720016477 Barthel T, Huschmann FU, Wallacher D, Feiler CG, Klebe G, Weiss MS, Wollenhaupt J. J. Appl. Cryst. 2021, 54:376-382 |
 |
|
15 |
Oct 23, 2020 |
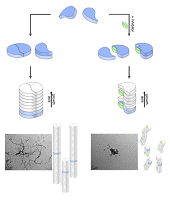
doi: 10.7554/eLife.59306 Lieblein T, Zangl R, Martin J, Hoffmann J, Hutchison MJ, Stark T, Stirnal E, Schrader T, Schwalbe H, Morgner N. eLife 2020;9:e59306 |
 |
|
14 |
Dec 14, 2020 |
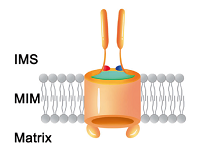
doi: 10.1038/s41422-020-00452-y Zhou S, Ruan M, Li Y, Yang J, Bai S, Richter C, Schwalbe H, Xie C, Shen B, Wang J. Cell Research, 2020, 0:1-4 |
 |
|
13 |
Dec 10, 2021 |
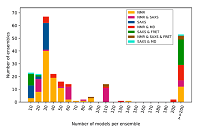
PED in 2021: a major update of the protein ensemble database for intrinsically disordered proteins doi: 10.1093/nar/gkaa1021 Lazar T, Martínez-Pérez E, Quaglia F, Hatos A, Chemes LB, Iserte JA, Méndez NA, Garrone NA, Saldaño TE, Marchetti J, Velez Rueda AJ, Bernadó P, Blackledge M, Cordeiro TN, Fagerberg E, Forman-Kay JD, Fornasari MS, Gibson TJ, Gomes GNW, Gradinaru CC, Head-Gordon T, Ringkjøbing Jensen M, Lemke EA, Longhi S, Marino-Buslje C, Minervini G, Mittag T, Monzon AM, Pappu RV, Parisi G, Ricard-Blum S, Ruff KM, Salladini E, Skepö M, Svergun D, Vallet SD, Varadi M, Tompa P, Tosatto SCE, Piovesan D. Nucl. Acids Res. 2021, 49:D404-D411 |
 |
|
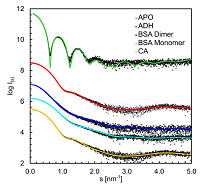
12 |
Oct 27, 2020 |
doi: 10.3390/cryst10110975 Graewert MA, Da Vela S, Graewert TW, Molodenskyi DS, Blanchet CE, Svergun DI, Jeffreis CM. Crystals 2020, 10:975 |
 |
|
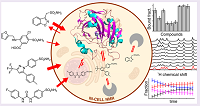
11 |
Sep 21, 2021 |
doi: 10.1021/acschembio.0c00590 Luchinat E, Barbieri L, Cremonini M, Nocentini A, Supuran CT, Banci L. ACS Chem. Biol. 2020, 15:2792-2800 |
 |
|
10 |
Jun 18, 2021 |
doi: https://dx.doi.org/10.1021/acs.analchem.0c01677 Luchinat E, Barbieri L, Campbell TF, Banci L. Anal. Chem. 2020, 92:9997-10006 |
 |
|
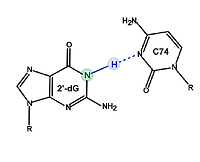
9 |
Sep 25, 2020 |
In-Cell NMR Spectroscopy of Functional Riboswitch Aptamers in Eukaryotic Cells doi: 10.1002/anie.202007184 Broft P, Dzatko S, Krafcikova M, Wacker A, Hänsel-Hertsch R, Dötsch V, Trantirek L, Schwalbe H. Angew. Chem. Int. Ed. 2021, 60:865-872 |
 |
|
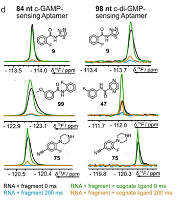
8 |
Sep 25, 2020 |
doi: 10.1002/cbic.202000476 Binas O, de Jesus V, Landgraf T, Völklein AE, Martins J, Hymon D, Kaur Bains J, Berg H, Biedenbänder T, Fürtig B, Lakshmi Gande S, Niesteruk A, Oxenfarth A, Shahin Qureshi N, Schamber T, Schnieders R, Tröster A, Wacker A, Wirmer-Bartoschek J, Wirtz Martin MA, Stirnal E, Azzaoui K, Richter C, Sreeramulu S, José Blommers MJ, Schwalbe H. ChemBioChem 2021, 22:423-433 |
 |
|
7 |
Jul 22, 2020 |
Anomeric Selectivity of Trehalose Transferase with Rare L‑Sugars doi: 10.1021/acscatal.0c02117 Mestrom L, Marsden SR, van der Eijk H, Laustsen JU, Jeffries CM, Svergun DI, Hagedoorn PL, Bento I, Hanefeld U. ACS Catal. 2020, 10:8835-8839 |
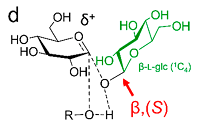 |
|
6 |
Jul 16, 2020 |
Multitarget virtual screening for drug repurposing in COVID19 doi: 10.26434/chemrxiv.12652997.v2 Sorzano CO, Crisman E, Carazo JM, Leon R. ChemRxiv 2020 |
 |
|
5 |
Jun 23, 2020 |
doi: 10.1016/j.celrep.2020.107817 Burda PC, Crosskey T, Lauk K, Zurborg A, SOhnchen C, Liffner B, Wilcke L, Pietsch E, Strauss J, Jeffries CM, Svergun DI, Wilson DW, Wilmanns M, Gilberger TW. Cell Reports 2020, 31:107817 |
 |
|
4 |
Jul 1, 2020 |
The basics of small-angle neutron scattering (SANS for new users of structural biology) doi: 10.1051/epjconf/202023603001 Jeffries CM, Pietras Z, Svergun DI. EPJ Web of Conferences 2020, 236:03001 |
 |
|
3 |
Jun 19, 2020 |
Development of in vitro‑grown spheroids as a 3D tumor model system for solid‑state NMR spectroscopy doi: 10.1007/s10858-020-00328-8 Damman R, Lucini Paloni A, Xenaki KT, Hernandez IB Van Bergen en Henegouwen PMP, Baldus M. J. Biomol. NMR 2020, 74:401-412. |
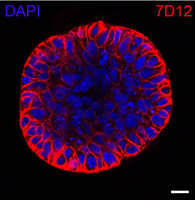 |
|
2 |
Sep 17, 2020 |
Improvements on marker-free images alignment for electron tomography doi: 10.1016/j.yjsbx.2020.100037 Sorzano COS, de Isidro-Gómez F, Fernández-Giménez E, Herreros D, Marco S, Carazo JM, Messaoudi C. J. Struct. Biol. X. 2020, 4:100037 |
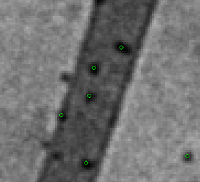 |
|
1 |
Jun 12, 2020 |
NMR quality control of fragment libraries for screening doi: 10.1007/s10858-020-00327-9 Sreeramulu S, Richter C, Kuehn T, Azzaoui K, Blommers MJJ, Del Conte R, Fragai M, Trieloff N, Schmieder P, Nazaré M, Specker E, Ivanov V, Oschkinat H, Banci L, Schwalbe H. J. Biomol. NMR. 2020, 74(10-11):555-563 |
 |
< 21-40