All publications from iNEXT-Discovery acknowledge the project and are appearing as Open Access!
|
40 |
March 19, 2021 |
Strategies for Optimization of Cryogenic Electron Tomography Data Acquisition doi: 10.3791/62383 Weis F, Hagen WJH, Schorb M, Mattei S. J Vis Exp.; 2021, Mar 19:169 |
 |
|
39 |
June 17, 2021 |
A Sample Preparation Pipeline for Microcrystals at the VMXm Beamline doi: 10.3791/62306 Crawshaw AD, Beale EV, Warren AJ, Stallwood A, Duller G, Trincao J, Evans G. J Vis Exp.; 2021, Jun 17:172 |
 |
|
38 |
May 28, 2021 |
Cryo-Structured Illumination Microscopic Data Collection from Cryogenically Preserved Cells doi: 10.3791/62274 Vyas N, Perry N, Okolo CA, Kounatidis I, Fish TM, Nahas KL, Jadhav A, Koronfel MA, Groen J, Pereiro E, Dobbie IM, Harkiolaki M. J Vis Exp.; 2021, May 28:171 |
 |
|
37 |
June 4, 2021 |
NMR-based Fragment Screening in a Minimum Sample but Maximum Automation Mode doi: 10.3791/62262 Berg H, Wirtz Martin MA, Niesteruk A, Richter C, Sreeramulu S, Schwalbe H. J Vis Exp.; 2021, Jun 4:172 |
 |
|
36 |
May 29, 2021 |
Cryo-EM and Single-Particle Analysis with Scipion doi: 10.3791/62261 Jiménez-Moreno A, del Caño L, Martínez M, Ramírez-Aportela E, Cuervo A, Melero R, Sánchez-García R, Strelak D, Fernández-Giménez E, de Isidro-Gómez F, Herreros D, Conesa P, Fonseca Y, Maluenda D, Jiménez de la Morena J, Macías J, Losana P, Marabini R, Carazo J, Sorzano C. J Vis Exp.; 2021, May 29:171 |
 |
|
35 |
March 3, 2021 |
Workflow and Tools for Crystallographic Fragment Screening at the Helmholtz-Zentrum Berlin doi: 10.3791/62208 Wollenhaupt J, Barthel T, Lima GMA, Metz A, Wallacher D, Jagudin E, Huschmann FU, Hauß T, Feiler CG, Gerlach M, Hellmig M, Förster R, Steffien M, Heine A, Klebe G, Mueller U, Weiss, MS. J Vis Exp.; 2021, Mar 3:169 |
 |
|
34 |
March 15, 2021 |
A 3D Cartographic Description of the Cell by Cryo Soft Xray Tomography doi: 10.3791/62190 Groen J, Sorrentino A, Aballe L, Oliete R, Valcarcel R, Okolo C, Kounatidis I Harkiolaki M, Perez-Berna AJ, Pereiro E. J Vis Exp.; 2021, Mar 15:169 |
 |
|
33 |
May 16, 2021 |
Nano-Differential Scanning Fluorimetry for Screening in Fragment-based Lead Discovery doi: 10.3791/62469 Ahmad MUD, Fish, A, Molenaar J, Sreeramulu S, Richter C, Altincekic N, Schwalbe H, Wienk H, Perrakis A. J Vis Exp.; 2021, May 16:171 |
 |
|
32 |
May 14, 2021 |
doi: 10.1038/s41596-021-00522-4 Okolo CH, Kounatidis I, Groen J, Nahas KL, Balint S, Fish TM Koronfel MA, Cortajarena AL, Dobbie IM, Pereiro E, Harkiolaki M. Nat Protoc. 2021;16(6):2851-2885 |
 |
|
31 |
February 12, 2021 |
Modelling covalent linkages in CCP4 doi: 10.1107/S2059798321001753 Nicholls RA, Joosten RP, Long F, Wojdyr M, Lebedev A, Krissinel E, Catapano L, Fischer M, Emsley P, Murshudov GN. Acta Crystallogr D Struct Biol. 2021;77(Pt 6):712-726 |
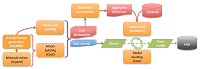 |
|
30 |
April 13, 2021 |
The missing link: covalent linkages in structural models doi: 10.1107/S2059798321003934 Nicholls RA, Wojdyr M, Joosten RP, Catapano L, Long F, Fischer M, Emsley P, Murshudov GN. Acta Crystallogr D Struct Biol. 2021;77(Pt 6):727-745 |
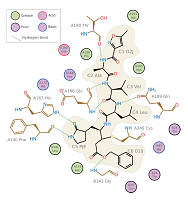 |
|
29 |
May 10, 2021 |
Large-Scale Recombinant Production of the SARS-CoV-2 Proteome for High-Throughput and Structural doi: 10.3389/fmolb.2021.653148 Altincekic N, Korn SM, Qureshi NS, Dujardin M, Ninot-Pedrosa M, Abele R, Abi Saad MJ, Alfano C, Almeida FCL, Alshamleh I, de Amorim GC, Anderson TK, Anobom CD, Anorma C, Bains JK, Bax A, Blackledge M, Blechar J, Böckmann A, Brigandat L, Bula A, Bütikofer M, Camacho-Zarco AR, Carlomagno T, Caruso IP, Ceylan B, Chaikuad A, Chu F, Cole L, Crosby MG, de Jesus V, Dhamotharan K, Felli IC, Ferner J, Fleischmann Y, Fogeron M-L, Fourkiotis NK, Fuks C, Fürtig B, Gallo A, Gande SL, Gerez JA, Ghosh D, Gomes-Neto F, Gorbatyuk O, Guseva S, Hacker C, Häfner S, Hao B, Hargittay B, Henzler-Wildman K, Hoch JC, Hohmann KF, Hutchison MT, Jaudzems K, Jovic´ K, Kaderli J, Kalnin¸ sˇ G, Kan¸ epe I, Kirchdoerfer RN, Kirkpatrick J, Knapp S, Krishnathas R, Kutz F, zur Lage S, Lambertz R, Lang A, Laurents D, Lecoq L, Linhard V, Löhr F, Malki A, Bessa LM, Martin RW, Matzel T, Maurin D, McNutt SW, Mebus-Antunes NC, Meier BH, Meiser N, Mompeán M, Monaca E, Montserret R, Mariño Perez L, Moser C, Muhle-Goll C, Neves-Martins TC, Ni X, Norton-Baker B, Pierattelli R, Pontoriero L, Pustovalova Y, Ohlenschläger O, Orts J, Da Poian AT, Pyper DJ, Richter C, Riek R, Rienstra CM, Robertson A, Pinheiro AS, Sabbatella R, Salvi N, Saxena K, Schulte L, Schiavina M, Schwalbe H, Silber M, Almeida MdS, Sprague-Piercy MA, Spyroulias GA, Sreeramulu S, Tants J-N, Tars K, Torres F, Töws S, Treviño MÁ, Trucks S, Tsika AC, Varga K, Wang Y, Weber ME, Weigand JE, Wiedemann C,Wirmer-Bartoschek J, Wirtz Martin MA, Zehnder J, Hengesbach M and Schlundt A. Front. Mol. Biosci. 2021, 8:653148 |
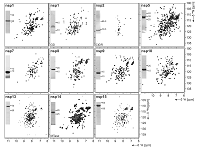 |
|
28 |
Jun 18, 2021 |
doi: 10.1016/j.xpro.2021.100529 Vyas N, Kunne S, Fish TM Dobbie IA, Harkiolaki M, Paul-Gilloteaux P. STAR Protocols 2021, 2:100529 |
 |
|
27 |
Apr 8, 2021 |
FragMAXapp: crystallographic fragment-screening data-analysis and project-management system doi: 10.1107/S2059798321003818 Lima GMA, Jagudin E, Talibov VO, Benz LS, Marullo C, Barthel T, Wollenhaupt J, Weiss MS, Mueller U. Acta Cryst. 2021, D77:799–808 |
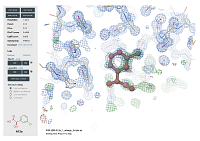 |
|
26 |
Mar 30, 2021 |
doi: 10.1021/jacs.1c01914 Kim J, Novakovic M, Jayanthi S, Lupulescu A, Kupce E, Grun JT, Mertinkus K, Oxenfarth A, Richter C, Schnieders R, Wirmer-Bartoschek J, Schwalbe H, Frydman L. J. Am. Chem. Soc. 2021, 143:4942−4948 |
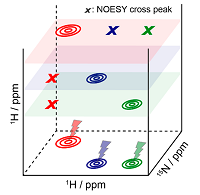 |
|
25 |
Mar 13, 2021 |
doi: 10.1007/s12104-021-10019-6 Wang Y, Kirkpatrick J, Zur Lage S, Korn SM, Neißner K, Schwalbe H, Schlundt A, Carlomagno T. Biomol NMR Assign.2021, 15:287-295 |
 |
|
24 |
Mar 29, 2021 |
doi: 10.1093/nar/gkab184 Marcelot A, Petitalot A, Ropars V, Le Du M-H, Samson C, Dubois S, Hoffmann G, Miron S, Cuniasse P, Marquez JA, Thai R, Theillet F-X, Zinn-Justin S. Nucl. Acid Res. 2021, 49(7):3841-3855 |
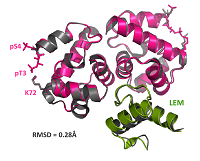 |
|
23 |
Mar 9, 2021 |
doi: 10.3791/62323 Barbieri L, Luchinat E. J. Vis. Exp. 2021, 169:e62323 |
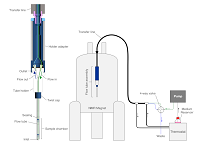 |
|
22 |
May 6, 2021 |
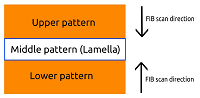
Cryo-focused ion beam lamella preparation protocol for in situ Structural Biology doi: 10.1007/978-1-0716-1406-8 Moravcová J, Dopitová R, Pinkas M, Nováček J. Methods Mol Biol. 2021, 2305:301-322 |
 |
|
21 |
Mar 8, 2021 |
doi: 10.1002/anie.202015948 Novakovic M, Kupce E, Scherf T, Oxenfarth A, Schnieders R, Grun JT, Wirmer-Bartoschek J, Richter C, Schwalbe H, Frydman L. Angew. Chem. Int. Ed. 2021, 60:2–10 |
 |