All publications from iNEXT-Discovery acknowledge the project and appear as Open Access!
|
240 |
Jan 07, 2025 |
doi: 10.1093/nar/gkae1235 Mortensen S, Kuncová S, Lormand JD, Myers TM, Kim SK, Lee VT, Winkler WC, Sondermann H. Nucleic Acids Res. 2025 Jan 7;53(1):gkae1235 |
 |
|
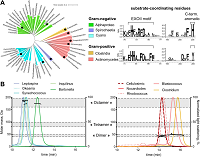
239 |
Jan 07, 2025 |
Magnetotactic bacteria from diverse Pseudomonadota families biomineralize intracellular Ca-carbonate doi: 10.1093/ismejo/wrae260 Mangin CC, Benzerara K, Bergot M, Menguy N, Alonso B, Fouteau S, Méheust R, Chevrier D, Godon C, Turrini E, Mehta N, Duverger A, Travert C, Busigny V, Duprat E, Bolzoni R, Cruaud C, Viollier E, Jézéquel D, Vallenet D, Lefèvre CT, Monteil CL. ISME J. 2025 Jan 7:wrae260 |
 |
|
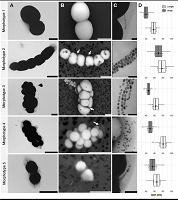
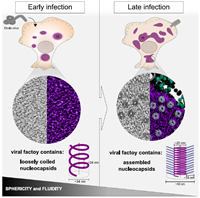
238 |
Feb 06, 2025 |
Nucleocapsid assembly drives Ebola viral factory maturation and dispersion doi: 10.1016/j.cell.2024.11.024 Vallbracht M, Bodmer BS, Fischer K, Makroczyova J, Winter SL, Wendt L, Wachsmuth-Melm M, Hoenen T, Chlanda P. Cell. 2024 Dec 14:S0092-8674(24)01340-0 |
 |
|
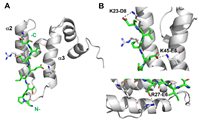
237 |
Dec 24, 2024 |
NMR Studies on the Structure of Yeast Sis1 and the Dynamics of doi: 10.3390/molecules30010011 Matos CO, Pinheir GMS, Caruso IP, Amorim GC, Almeida FCL, Ramos CHI. Molecules 2025, 30, 11 |
 |
|
236 |
Nov 07, 2024 |
Cryo-EM structure of a lysozyme-derived amyloid fibril from hereditary amyloidosis doi: 10.1038/s41467-024-54091-7 Karimi-Farsijani S, Sharma K, Ugrina M, Kuhn L, Pfeiffer PB, Haupt C, Wiese S, Hegenbart U, Schönland SO, Schwierz N, Schmidt M, Fändrich M. Nat Commun 15, 9648 (2024) |
 |
|
235 |
Oct 18, 2024 |
doi: 10.1038/s44318-024-00264-5 Rosa LT, Vernhes É, Soulet AL, Polard P, Fronzes R. EMBO J. 2024 Oct 18. Epub ahead of print. |
 |
|
234 |
Oct 09, 2024 |
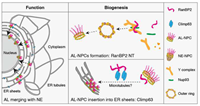
Annulate Lamellae biogenesis is essential for nuclear pore function doi: 10.21203/rs.3.rs-5104543 Lin J, Agote-Arán A, Lia Y, Schoch R, Ronchi P, Cochard V, Zhu R, Kleiss C, Ruff M, Chevreux G, Schwab Y, Klaholz BP, Sumara I. Preprint (Version 1); Research Square |
 |
|
233 |
Oct 01, 2024 |
doi: 10.1107/S2059798324009252 Sotiropoulou AI, Hatzinikolaou DG, Chrysina ED. Acta Crystallogr D Struct Biol. 2024 Oct 1;80(Pt 10):733-743 |
 |
|
232 |
Oct 20, 2022 |
doi: 10.1107/S2059798322009184 Trewhella J, Vachette P, Bierma J, Blanchet C, Brookes E, Chakravarthy S, Chatzimagas L, Cleveland TE 4th, Cowieson N, Crossett B, Duff AP, Franke D, Gabel F, Gillilan RE, Graewert M, Grishaev A, Guss JM, Hammel M, Hopkins J, Huang Q, Hub JS, Hura GL, Irving TC, Jeffries CM, Jeong C, Kirby N, Krueger S, Martel A, Matsui T, Li N, Pérez J, Porcar L, Prangé T, Rajkovic I, Rocco M, Rosenberg DJ, Ryan TM, Seifert S, Sekiguchi H, Svergun D, Teixeira S, Thureau A, Weiss TM, Whitten AE, Wood K, Zuo X. Acta Crystallogr D Struct Biol. 2022 Nov 1;78(Pt 11):1315-1336 |
 |
|
231 |
Sep 19, 2024 |
Kinetic and Structural Studies of the Plastidial Solanum tuberosum doi: 10.1021/acsomega.4c06313 Koulas SM, Kyriakis E, Tsagkarakou AS, Leonidas DD. ACS Omega 2024 |
 |
|
230 |
Jan 11, 2023 |
Estimating conformational landscapes from Cryo-EM particles by 3D Zernike polynomials doi: 10.1038/s41467-023-35791-y Herreros D, Lederman RR, Krieger JM, Jiménez-Moreno A, Martínez M, Myška D, Strelak D, Filipovic J, Sorzano COS, Carazo JM. Nat Commun. 2023 Jan 11;14(1):154 |
 |
|
229 |
Aug 01, 2023 |
doi: 10.1002/anie.202303202 Cerofolini L, Vasa K, Bianconi E, Salobehaj M, Cappelli G, Bonciani A, Licciardi G, Pérez-Ràfols A, Padilla-Cortés L, Antonacci S, Rizzo D, Ravera E, Viglianisi C, Calderone V, Parigi G, Luchinat C, Macchiarulo A, Menichetti S, Fragai M. Angew Chem Int Ed Engl. 2023 Aug 1;62(31):e202303202 |
 |
|
228 |
Sep 18, 2023 |
Effects of boron supplementation on dairy calves' health: a metabolomics study doi: 10.21608/avmj.2023.218730.1157 Adam A, Baspinar N, Vignoli A, Meoni G, Tenori L, Basoglu A, Gulersoy E, Bicici R. Assiut Vet. Med. J. Vol. 70 No. 180 January 2024, 10-25 |
 |
|
227 |
Apr 03, 2023 |
Modulation of Aβ42 Aggregation Kinetics and Pathway by Low-Molecular-Weight Inhibitors doi: 10.1002/cbic.202200760 Hutchison MT, Bellomo G, Cherepanov A, Stirnal E, Fürtig B, Richter C, Linhard V, Gurewitsch E, Lelli M, Morgner N, Schrader T, Schwalbe H. Chembiochem. 2023 Apr 3;24(7):e202200760 |
 |
|
226 |
Apr 25, 2023 |
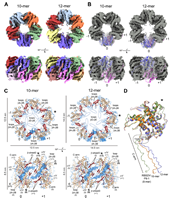
doi: 10.1128/mbio.00023-23 Llauger G, Melero R, Monti D, Sycz G, Huck-Iriart C, Cerutti ML, Klinke S, Mikkelsen E, Tijman A, Arranz R, Alfonso V, Arellano SM, Goldbaum FA, Sterckx YGJ, Carazo JM, Kaufman SB, Dans PD, Del Vas M, Otero LH. mBio. 2023 Apr 25;14(2):e0002323 |
 |
|
225 |
Mar 10, 2023 |
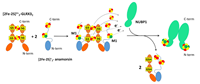
doi: 10.1002/pro.4625 Bargagna B, Matteucci S, Ciofi-Baffoni S, Camponeschi F, Banci L. Protein Sci. 2023 May;32(5):e4625 |
 |
|
224 |
Apr 19, 2024 |
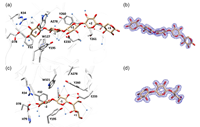
doi: 10.1002/bit.28731 Pentari C, Kosinas C, Nikolaivits E, Dimarogona M, Topakas E. Biotechnol Bioeng. 2024 Jul;121(7):2067-2078 |
 |
|
223 |
Jan 09, 2024 |
Controlling the incorporation of fluorinated amino acids in human cells and its structural impact doi: 10.1002/pro.4910 Costantino A, Pham LBT, Barbieri L, Calderone V, Ben-Nissan G, Sharon M, Banci L, Luchinat E. Protein Sci. 2024 Mar;33(3):e4910 |
 |
|
222 |
Apr 19, 2024 |
doi: 10.7554/eLife.93759 Petit-Hartlein I, Vermot A, Thepaut M, Humm AS, Dupeux F, Dupuy J, Chaptal V, Marquez JA, Smith SME, Fieschi F. Elife. 2024 Apr 19;13:RP93759 |
 |
|
221 |
Jul 23, 2024 |
Controlled pH alteration enables guanine accumulation and drives crystallization within iridosomes doi: 10.1101/2024.07.20.604036 Eyal Z, Gorelick-Ashkenazi A, Deis R, Barzilay Y, Yonatan Broder Y, Perry Kellum A, Varsano N, Hartstein M, Sorrentino A, Kaplan-Ashiri I, Rechav K, Metzler R, Houben L, Kronik L, Rez P, Gur D. BioRxiv |
 |