All publications from iNEXT-Discovery acknowledge the project and appear as Open Access!
|
220 |
Aug 11, 2023 |
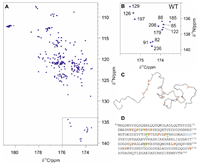
Studies of proline conformational dynamics in IDPs by 13C-detected cross-correlated NMR relaxation doi: 10.1016/j.jmr.2023.107539 Schiavina M, Konrat R, Ceccolini I, Mateos B, Konrat R, Felli IC, Pierattelli R. J Magn Reson. 2023 Sep;354:107539
|
 |
|
219 |
May 13, 2022 |
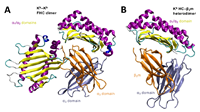
Dissociation of β2m from MHC class I triggers formation of noncovalent transient heavy chain dimers doi: 10.1242/jcs.259498 Dirscherl C, Löchte S, Hein Z, Kopicki JD, Harders AR, Linden N, Karner A, Preiner J, Weghuber J, Garcia-Alai M, Uetrecht C, Zacharias M, Piehler J, Lanzerstorfer P, Springer S. J Cell Sci. 2022 May 1;135(9):jcs259489 |
 |
|
218 |
Sep 18, 2024 |
Structural basis of CDNF interaction with the UPR regulator GRP78 doi: 10.1038/s41467-024-52478-0 Graewert MA, Volkova M, Jonasson K, Määttä JAE, Gräwert T, Mamidi S, Kulesskaya N, Evenäs J, Johnsson RE, Svergun D, Bhattacharjee A, Huttunen HJ. Nat Commun. 2024 Sep 18;15(1):8175 |
 |
|
217 |
Oct 03, 2024 |
The future of integrated structural biology doi: 10.1016/j.str.2024.08.014 Schwalbe H, Audergon P, Haley N, Alen Amaro C, Agirre J, Baldus M, Banci L, Baumeister W, Blackledge M, Carazo JM, Djinovic Carugo K, Celie P, Felli I, Hart DJ, Hauß T, Lehtiö L, Lindorff-Larsen K, Márquez J, Matagne A, Pierattelli R, Rosato A, Sobott F, Sreeramulu S, Steyaert J, Sussman JL, Trantirek L, Weiss MS, Wilmanns M. Structure (2024) |
 |
|
216 |
Sep 05, 2024 |
A bitopic agonist bound to the dopamine 3 receptor reveals a selectivity site doi: 10.1038/s41467-024-51993-4 Arroyo-Urea S, Nazarova AL, Carrión-Antolí Á, Bonifazi A, Battiti FO, Lam JH, Newman AH, Katritch V, García-Nafría J. Nat Commun. 2024 Sep 5;15(1):7759 |
 |
|
215 |
Aug 24, 2024 |
Structural and biochemical characterization of the C-terminal region of the human RTEL1 helicase doi: 10.1002/pro.5093 Cortone G, Graewert MA, Kanade M, Longo A, Hegde R, González-Magaña A, Chaves-Arquero B, Blanco FJ, Napolitano LMR, Onesti S. Protein Sci. 2024 Sep;33(9):e5093 |
 |
|
214 |
Aug 08, 2024 |
Optimising in-cell NMR acquisition for nucleic acids doi: 10.1007/s10858-024-00448-5 Annecke HTP, Eidelpes R, Feyrer H, Ilgen J, Gürdap CO, Dasgupta R, Petzold K. J Biomol NMR. 2024 Aug 20 |
 |
|
213 |
Aug 16, 2024 |
doi: 10.1093/nar/gkae704 Hernández-Marín M, Cantero-Camacho Á, Mena I, López-Núñez S, García-Sastre A, Gallego J. Nucleic Acids Res. 2024 Aug 16:gkae704 |
 |
|
212 |
Aug 08, 2024 |
An integrative structural study of the human full-length RAD52 at 2.2 Å resolution doi: 10.1038/s42003-024-06644-1 Balboni B, Marotta R, Rinaldi F, Milordini G, Varignani G, Girotto S, Cavalli A. Commun Biol. 2024 Aug 8;7(1):956 |
 |
|
211 |
Jun 26, 2024 |
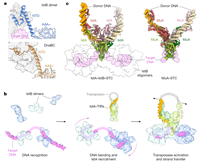
Molecular basis for transposase activation by a dedicated AAA+ ATPase doi: 10.1038/s41586-024-07550-6 de la Gándara Á, Spínola-Amilibia M, Araújo-Bazán L, Núñez-Ramírez R, Berger JM, Arias-Palomo E. Nature. 2024 Jun;630(8018):1003-1011 |
 |
|
210 |
Jun 06, 2024 |
doi: 10.1038/s41594-024-01274-x Holvec S, Barchet C, Lechner A, Fréchin L, De Silva SNT, Hazemann I, Wolff P, von Loeffelholz O, Klaholz BP. Nat Struct Mol Biol. 2024 Jun 6 |
 |
|
209 |
May 16, 2024 |
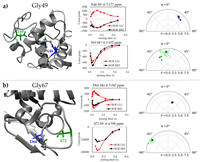
Improved detection of magnetic interactions in proteins based on long-lived coherences doi: 10.1038/s42004-024-01195-2 Ianc O, Teleanu F, Ciumeică A, Lupulescu A, Sadet A, Vasos PR. Commun Chem. 2024 May 16;7(1):112 |
 |
|
208 |
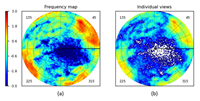
May 10, 2024 |
Program VUE: analysing distributions of cryo-EM projections using uniform spherical grids doi: 10.1107/s1600576724002383 Urzhumtseva L, Barchet C, Klaholz BP, Urzhumtsev AG. J. Appl. Cryst. 57 |
 |
|
207 |
Apr 15, 2024 |
Progression of herpesvirus infection remodels mitochondrial organization and metabolism doi: 10.1371/journal.ppat.1011829 Leclerc S, Gupta A, Ruokolainen V, Chen JH, Kunnas K, Ekman AA, Niskanen H, Belevich I, Vihinen H, Turkki P, Perez-Berna AJ, Kapishnikov S, Mäntylä E, Harkiolaki M, Dufour E, Hytönen V, Pereiro E, McEnroe T, Fahy K, Kaikkonen MU, Jokitalo E, Larabell CA, Weinhardt V, Mattola S, Aho V, Vihinen-Ranta M. PLoS Pathog. 2024 Apr 15;20(4):e1011829 |
 |
|
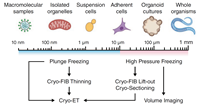
206 |
Apr 30, 2024 |
Cryogenic Preparations of Biological Specimens for Cryo-Electron Tomography doi: 10.1007/978-3-031-51171-4_3 D’Imprima E, Fung HKH, Zagoriy I, Mahamid J. In: Cryo-Electron Tomography. Focus on Structural Biology, vol 11. Springer, Cham. |
 |
|
205 |
Apr 23, 2024 |
UBAP2L ensures homeostasis of nuclear pore complexes at the intact nuclear envelope doi: 10.1083/jcb.202310006 Liao Y, Andronov L, Liu X, Lin J, Guerber L, Lu L, Agote-Arán A, Pangou E, Ran L, Kleiss C, Qu M, Schmucker S, Cirillo L, Zhang Z, Riveline D, Gotta M, Klaholz BP, Sumara I. J Cell Biol. 2024 Jul 1;223(7):e202310006 |
 |
|
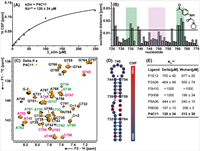
204 |
Apr 02, 2024 |
doi: 10.1261/rna.079902.123 Matzel T, Wirtz Martin M, Herr A, Wacker A, Richter C, Sreeramulu S, Schwalbe H. RNA. 2024 Apr 2:rna.079902.123 |
 |
|
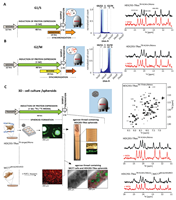
203 |
Mar 22, 2024 |
doi: 10.26434/chemrxiv-2024-jqd66-v2 Rynes J, Istvankova E, Krafcikova M, Luchinat E, Barbieri L, Banci L, Kamarytova K, Loja T, Fafilek B, Rico-Llanos G, Krejci P, Macurek L, Foldynova-Trantirkova S, Trantirek L. https://doi.org/10.26434/chemrxiv-2024-jqd66-v2 |
 |
|
202 |
Mar 19, 2024 |
doi: 10.1038/s41467-024-46761-3 Morici M, Gabrielli S, Fujiwara K, Paternoga H, Beckert B, Bock LV, Chiba S, Wilson DN. Nat Commun. 2024 Mar 19;15(1):2432 |
 |
|
201 |
Mar 05, 2024 |
In-cell NMR suggests that DNA i-motif levels are strongly depleted in living human cells doi: 10.1038/s41467-024-46221-y Víšková P, Ištvánková E, Ryneš J, Džatko Š, Loja T, Živković ML, Rigo R, El-Khoury R, Serrano-Chacón I, Damha MJ, González C, Mergny JL, Foldynová-Trantírková S, Trantírek L. Nat Commun. 2024 Mar 5;15(1):1992 |
 |