All publications from iNEXT-Discovery acknowledge the project and appear as Open Access!
|
180 |
Jul 04, 2023 |
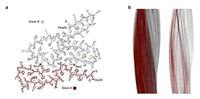
Structural analysis of human ex vivo amyloid fibrils from systemic AA amyloidosis Banerjee S (PhD Thesis) https://oparu.uni-ulm.de/xmlui/bitstream/handle/123456789/51345/Banerjee.pdf?sequence=1 |
 |
|
179 |
Oct 26, 2023 |
Crystallographic Fragment Screening on the Shigella Type III Secretion System Chaperone IpgC doi: 10.1021/acsomega.3c07058 Gárdonyi M, Hasewinkel C, Wallbaum J, Wollenhaupt J, Weiss MS, Klebe G, Reuter K, Heine A ACS Omega 2023 |
 |
|
178 |
Nov 03, 2023 |
Gulotta A, Polimeni M, Lenton S, Starr CG, Kingsbury JS, Stradner A, Zaccarelli E, Schurtenberger P. link: https://arxiv.org/pdf/2311.01986.pdf
|
 |
|
177 |
Oct 30, 2023 |
Site-Selective Functionalized PD-1 Mutant for a Modular Immunological Activity against Cancer Cells doi: 10.1021/acs.biomac.3c00893 Fallarini S, Cerofolini L, Salobehaj M, Rizzo D, Gheorghita GR, Licciardi G, Capialbi DE, Zullo V, Sodini A, Nativi C, Fragai M. Biomacromolecules |
 |
|
176 |
Oct 23, 2023 |
Staphylococcus aureus sacculus mediates activities of M23 hydrolases doi: 10.1038/s41467-023-42506-w Razew A, Laguri C, Vallet A, Bougault C, Kaus-Drobek M, Sabala I, Simorre JP. Nat Commun. 2023 Oct 23;14(1):6706 |
 |
|
175 |
Oct 20, 2023 |
Structure of the Borrelia bacteriophage φBB1 procapsid doi: 10.1016/j.jmb.2023.168323 Rūmnieks J, Füzik T, Tārs K. J Mol Biol. 2023 Oct 20:168323 |
 |
|
174 |
Oct 10, 2023 |
doi: 10.1038/s42003-023-05402-z Pierson E, De Pol F, Fillet M, Wouters J. Commun Biol. 2023 Oct 10;6(1):1024 |
 |
|
173 |
Oct 05, 2023 |
doi: 10.1021/acs.jafc.3c04532 Riboni N, Bianchi F, Mattarozzi M, Caldara M, Gullì M, Graziano S, Maestri E, Marmiroli N, Careri M. J Agric Food Chem. 2023 |
 |
|
172 |
Oct 04, 2023 |
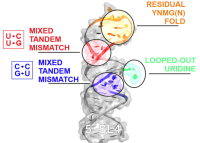
High-resolution structure of stem-loop 4 from the 5'-UTR of SARS-CoV-2 solved by solution state NMR doi: 10.1093/nar/gkad762 Vögele J, Hymon D, Martins J, Ferner J, Jonker HRA, Hargrove AE, Weigand JE, Wacker A, Schwalbe H, Wöhnert J, Duchardt-Ferner E. Nucleic Acids Res. 2023 Oct 4:gkad762 |
 |
|
171 |
Sep 21, 2023 |
Targeting EPHA2 with Kinase Inhibitors in Colorectal Cancer doi: 10.1002/cmdc.202300420 Tröster A, Jores N, Mineev KS, Sreeramulu S, DiPrima M, Tosato G, Schwalbe H. ChemMedChem. 2023 Sep 21:e202300420 |
 |
|
170 |
Oct 01, 2023 |
doi: 10.1107/S2059798323006514 Cavini IA, Winter AJ, D'Muniz Pereira H, Woolfson DN, Crump MP, Garratt RC. Acta Crystallogr D Struct Biol. 2023 |
 |
|
169 |
Sep 09, 2023 |
A thermosensitive gel matrix for bioreactor-assisted in-cell NMR of nucleic acids and proteins doi: 10.1007/s10858-023-00422-7 Dzurov M, Pospíšilová Š, Krafčíková M, Trantírek L, Vojtová L, Ryneš J. J Biomol NMR. 2023 |
 |
|
168 |
Sep 06, 2023 |
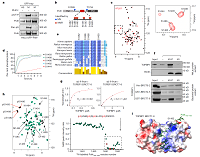
Polθ is phosphorylated by PLK1 to repair double-strand breaks in mitosis doi: 10.1038/s41586-023-06506-6 Gelot C, Kovacs MT, Miron S, Mylne E, Haan A, Boeffard-Dosierre L, Ghouil R, Popova T, Dingli F, Loew D, Guirouilh-Barbat J, Del Nery E, Zinn-Justin S, Ceccaldi R. Nature. 2023 Sep 6 |
 |
|
167 |
Sep 05, 2023 |
doi: 10.1039/d3sc02117c Beriashvili D, Yao R, D'Amico F, Krafčíková M, Gurinov A, Safeer A, Cai X, Mulder MPC, Liu Y, Folkers GE, Baldus M. Chem. Sci., 2023 |
 |
|
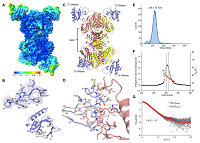
166 |
Aug 29, 2023 |
doi: 10.1016/j.celrep.2023.112972 Andriianov A, Trigüis S, Drobiazko A, Sierro N, Ivanov NV, Selmer M, Severinov K, Isaev A. Cell Rep. 2023;42(8):112972 |
 |
|
165 |
Aug 10, 2023 |
doi: 10.20944/preprints202308.0828.v1 Krieger JM, Sorzano COS, Carazo JM. Preprints 2023, 2023080828 |
 |
|
164 |
Aug 07, 2023 |
Structural conservation of antibiotic interaction with ribosomes doi: 10.1038/s41594-023-01047-y Paternoga H, Crowe-McAuliffe C, Bock LV, Koller TO, Morici M, Beckert B, Myasnikov AG, Grubmüller H, Nováček J, Wilson DN. Nat Struct Mol Biol. |
 |
|
163 |
Aug 04, 2023 |
doi: 10.3390/antib12030051 Meskova K, Martonova K, Hrasnova P, Sinska K, Skrabanova M, Fialova L, Njemoga S, Cehlar O, Parmar O, Kolenko P, Pevala V, Skrabana R. Antibodies 2023, 12, 51 |
 |
|
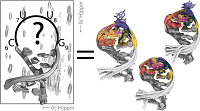
162 |
Jul 21, 2023 |
doi: 10.1021/jacs.3c03578 Oxenfarth A, Kümmerer F, Bottaro S, Schnieders R, Pinter G, Jonker HRA, Fürtig B, Richter C, Blackledge M, Lindorff-Larsen K, Schwalbe H. J Am Chem Soc. 2023;145(30):16557-16572 |
 |
|
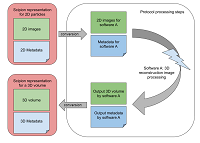
161 |
Jun 29, 2023 |
Scipion3: A workflow engine for cryo-electron microscopy image processing and structural biology doi: 10.1017/s2633903x23000132 Conesa P, Fonseca YC, Jiménez de la Morena J, Sharov G, de la Rosa-Trevín JM, Cuervo A, García Mena A, Rodríguez de Francisco B, del Hoyo D, Herreros D, Marchan D, Strelak D, Fernández-Giménez E, Ramírez-Aportela E, de Isidro-Gómez FP, Sánchez I, Krieger J, Vilas JL, del Cano L, Gragera M, Iceta M, Martínez M, Losana P, Melero R, Marabini R, Carazo JM, Sorzano COS. Biological Imaging, 1-22. |
 |