All publications from iNEXT-Discovery acknowledge the project and appear as Open Access!
|
200 |
Feb 26, 2024 |
A multi-scale numerical approach to study monoclonal antibodies in solution doi: 10.1063/5.0186642 Polimeni M, Zaccarelli E, Gulotta A, Lund M, Stradner A, Schurtenberger P. APL Bioengineering |
 |
|
199 |
Feb 15, 2024 |
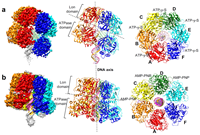
doi: 10.1101/2024.02.15.580443 Talachia Rosa L, Vernhes E, Soulet AL, Polard P, Fronzes R. bioRxiv preprint |
 |
|
198 |
Feb 14, 2024 |
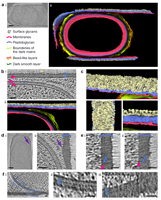
doi: 10.1038/s41467-024-45770-6 Bauda E, Gallet B, Moravcova J, Effantin G, Chan H, Novacek J, Jouneau PH, Rodrigues CDA, Schoehn G, Moriscot C, Morlot C. Nat Commun. 2024 Feb 14;15(1):1376 |
 |
|
197 |
Feb 12, 2024 |
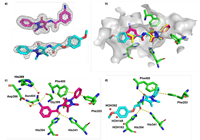
doi: 10.1039/d3md00724c Jalencas X, Berg H, Espeland LO, Sreeramulu S, Kinnen F, Richter C, Georgiou C, Yadrykhinsky V, Specker E, Jaudzems K, Mileti T, Harmel R, Gribbon P, Schwalbe H, Brenk R, Jirgensons A, Zaliani A, Mestres J. RSC Med. Chem., 2024 |
 |
|
196 |
Feb 01, 2024 |
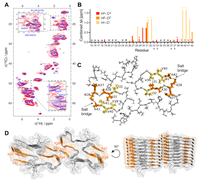
doi: 10.1038/s41467-024-45192-4 Kumar R, Le Marchand T, Adam L, Bobrovs R, Chen G, Fridmanis J, Kronqvist N, Biverstål H, Jaudzems K, Johansson J, Pintacuda G, Abelein A. Nat Commun. 2024 Feb 1;15(1):965 |
 |
|
195 |
Feb 06, 2024 |
doi: 10.1128/aac.00061-23 Brunetti F, Ghiglione B, Gudeta DD, Gutkind G, Guardabassi L, Klinke S, Power P. Antimicrob Agents Chemother. 2023 Jul 18;67(7):e0006123 |
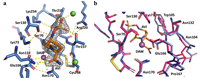 |
|
194 |
Jan 30, 2024 |
Improved detection of magnetic interactions in proteins based on long-lived coherences doi: 10.21203/rs.3.rs-3694602/v1 Sadet A, Ianc O, Teleanu F, Ciumeică A, Lupulescu A, Vasos P. Research Square, Version 1 |
 |
|
193 |
Jan 24, 2024 |
Molecular mechanisms and evolutionary robustness of a color switch in proteorhodopsins doi: 10.1126/sciadv.adj0384 Mao J, Jin X, Shi M, Heidenreich D, Brown LJ, Brown RCD, Lelli M, He X, Glaubitz C. Sci Adv. 2024 Jan 26;10(4):eadj0384 |
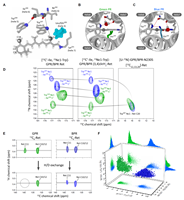 |
|
192 |
Jan 20, 2024 |
EasyGrid: A versatile platform for automated cryo-EM sample preparation and quality control doi: 10.1101/2024.01.18.576170 Gemin O, Armijo V, Hons M, Bissardon C, Linares R, Bowler MW, Wolff G, Kovalev K, Babenko A, Salo VT, Schneider S, Rossi C, Lecomte L, Deckers T, Lauzier K, Janocha R, Felisaz F, Sinoir J, Galej WP, Mahamid J, Muller CW, Eustermann S, Mattei S, Cipriani F, Papp G. bioRxiv preprint |
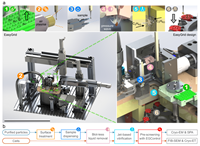 |
|
191 |
Jan 17, 2024 |
doi: 10.1021/acschembio.3c00720 Altincekic N, Jores N, Löhr F, Richter C, Ehrhardt C, Blommers MJJ, Berg H, Öztürk S, Gande SL, Linhard V, Orts J, Abi Saad MJ, Bütikofer M, Kaderli J, Karlsson BG, Brath U, Hedenström M, Gröbner G, Sauer UH, Perrakis A, Langer J, Banci L, Cantini F, Fragai M, Grifagni D, Barthel T, Wollenhaupt J, Weiss MS, Robertson A, Bax A, Sreeramulu S, Schwalbe H. ACS Chem Biol. 2024 Jan 17 |
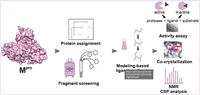 |
|
190 |
Jan 12, 2024 |
Ligand-Based Competition Binding by Real-Time 19F NMR in Human Cells doi: 10.1021/acs.jmedchem.3c01600 Luchinat E, Barbieri L, Davis B, Brough PA, Pennestri M, Banci L. J Med Chem. 2024 |
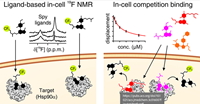 |
|
189 |
Jan 11, 2024 |
doi: 10.3390/molecules29020376 Wan Z, Shi M, Gong Y, Lucci M, Li J, Zhou J, Yang X-L, Lelli M, He X, Mao J. Molecules. 2024; 29(2):376 |
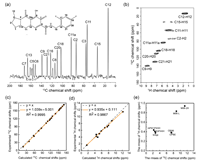 |
|
188 |
Jan 03, 2024 |
TIR domains produce histidine-ADPR conjugates as immune signaling molecules in bacteria doi: 10.1101/2024.01.03.573942 Sabonis D, Avraham C, Lu A, Herbst E, Silanskas A, Leavitt A, Yirmiya E, Zaremba M, Amitai G, Kranzusch PJ, Sorek R, Tamulaitiene G. bioRxiv preprint |
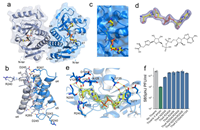 |
|
187 |
Dec 25, 2023 |
doi: 10.1002/advs.202306272 Maity A, Wulffelé J, Ayala I, Favier A, Adam V, Bourgeois D, Brutscher B. Adv Sci (Weinh). 2023 Dec 25:e2306272 |
 |
|
186 |
Dec 18, 2023 |
doi: 10.21203/rs.3.rs-3433207/v1 Arroyo-Urea S, Nazarova AL, Carrión-Antolí A, Bonifazi A, Battiti FO, Homing Lam J, Hauck Newman A, Katritch V, García-Nafría J. PREPRINT V1, available at Research Square |
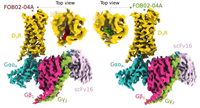 |
|
185 |
Dec 19, 2023 |
DNA i-motif levels are overwhelmingly depleted in living human cells: insights from in-cell NMR doi: 10.21203/rs.3.rs-3734993/v1 Trantirek L, Viskova P, Istvankova E, Rynes J, Dzatko S, Loja T, Lenarcic Zivkovic M, Rigo R, El-Khoury R, Serano I, Damha M, Gonzalez C, Mergny JL, Foldynova-Trantirkova S. PREPRINT V1, available at Research Square |
 |
|
184 |
Dec 13, 2023 |
Structural mechanisms of autoinhibition and substrate recognition by the ubiquitin ligase HACE1 doi: 10.21203/rs.3.rs-3220888/v1 Düring J, Wolter M, Toplak J, Torres C, Dybkov O, Fokkens TJ, Urlaub H, Steinchen W, Dienemann C, Lorenz S. PREPRINT V1, available at Research Square |
 |
|
183 |
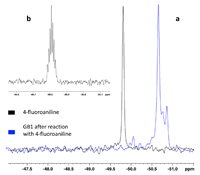
Dec 12, 2023 |
Enlarging the scenario of site directed 19F labeling for NMR spectroscopy of biomolecules doi: 10.1038/s41598-023-49247-2 Vitali V, Torricella F, Massai L, Messori L, Banci L. Sci Rep. 2023 Dec 12;13(1):22017 |
 |
|
182 |
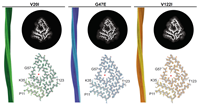
Nov 30, 2023 |
Kravikass M, Koren G, Saleh OA, Beck R. https://arxiv.org/pdf/2311.18337.pdf |
 |
|
181 |
Nov 22, 2023 |
Common transthyretin-derived amyloid fibril structures in patients with hereditary ATTR amyloidosis doi: 10.1038/s41467-023-43301-3 Steinebrei M, Baur J, Pradhan A, Kupfer N, Wiese S, Hegenbart U, Schönland SO, Schmidt M, Fändrich M. Nat Commun. 2023 Nov 22;14(1):7623 |
 |